One more Positron-related post! It’s a quick one, just to highlight one feature I think is really neat and helpful: the Connections Pane.
In a newer research project I’m working on, I have geocoded data for every foreign aid project run by most donor countries since 1989. This involves millions of rows and nearly a thousand columns across three CSV files, totalling 3.3 GB. R can load this data, but it takes a long time and slows down the session substantially (and often crashes R entirely).
Fortunately there’s a good solution for this huge data. DuckDB is a lightning fast database system that works really well with R. The DuckDB file that I’ve made for the foreign aid data compresses things substantially—it only takes up 600 MB on the disk instead of 3.3 GB. It also lets me query the huge data using either SQL commands or with {dplyr} functions, and I can load subsets of the huge data into R really quickly.
DuckDB, {DBI}, and the difficulty of discerning data in a database
Here’s a quick little example showing the general process with some toy data. I’ll create a little .duckdb file on the disk named example_database.duckdb—I could also make a temporary in-memory database by naming the file ":memory:":
Next I’ll add some stuff to it:
# Add stuff to the database
copy_to(con, penguins, name = "penguins", overwrite = TRUE, temporary = FALSE)
copy_to(con, gapminder::gapminder, name = "gapminder", overwrite = TRUE, temporary = FALSE)
copy_to(con, ggplot2::mpg, name = "mpg", overwrite = TRUE, temporary = FALSE)
copy_to(con, ggplot2::diamonds, name = "diamonds", overwrite = TRUE, temporary = FALSE)And then I’ll close the connection:
DBI::dbDisconnect(con)That worked, but it’s really hard to see if it did anything. The database file does contain stuff—it’s 1.6 MB now:
❯ ls -lh example_database.duckdb
# Permissions Size User Date Modified Name
# .rw-r--r--@ 1.6M andrew 2025-07-10 12:13 example_database.duckdb…and I can connect to the database with R and get data out of it:
con <- DBI::dbConnect(duckdb::duckdb(), db_file)
# Get stuff out of the database
adelie_query <- tbl(con, I("penguins")) |>
filter(species == "Adelie")
# Check it out! It's SQL!
show_query(adelie_query)
## <SQL>
## SELECT q01.*
## FROM (FROM penguins) q01
## WHERE (species = 'Adelie')
# Actually run the query
penguins_from_db <- adelie_query |>
collect()
penguins_from_db
## # A tibble: 152 × 8
## species island bill_len bill_dep flipper_len body_mass sex year
## <fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
## 1 Adelie Torgersen 39.1 18.7 181 3750 male 2007
## 2 Adelie Torgersen 39.5 17.4 186 3800 female 2007
## 3 Adelie Torgersen 40.3 18 195 3250 female 2007
## 4 Adelie Torgersen NA NA NA NA <NA> 2007
## 5 Adelie Torgersen 36.7 19.3 193 3450 female 2007
## 6 Adelie Torgersen 39.3 20.6 190 3650 male 2007
## 7 Adelie Torgersen 38.9 17.8 181 3625 female 2007
## 8 Adelie Torgersen 39.2 19.6 195 4675 male 2007
## 9 Adelie Torgersen 34.1 18.1 193 3475 <NA> 2007
## 10 Adelie Torgersen 42 20.2 190 4250 <NA> 2007
## # ℹ 142 more rows
DBI::dbDisconnect(con)…but there’s not an easy way to check what’s actually going on in there. I only know there’s a table named penguins in there because I just made it.
DuckDB provides a helpful browser-based UI that you can load from the terminal like this:
❯ duckdb -ui example_database.duckdbThis gives you a notebook where you can write SQL commands, which I find less helpful because I typically interact with the database with {dplyr}—I really only use DuckDB for fast data storage and fast data access. Most importantly, though, it gives you a sidebar that shows all the tables inside the database, and you can preview the stuff inside:
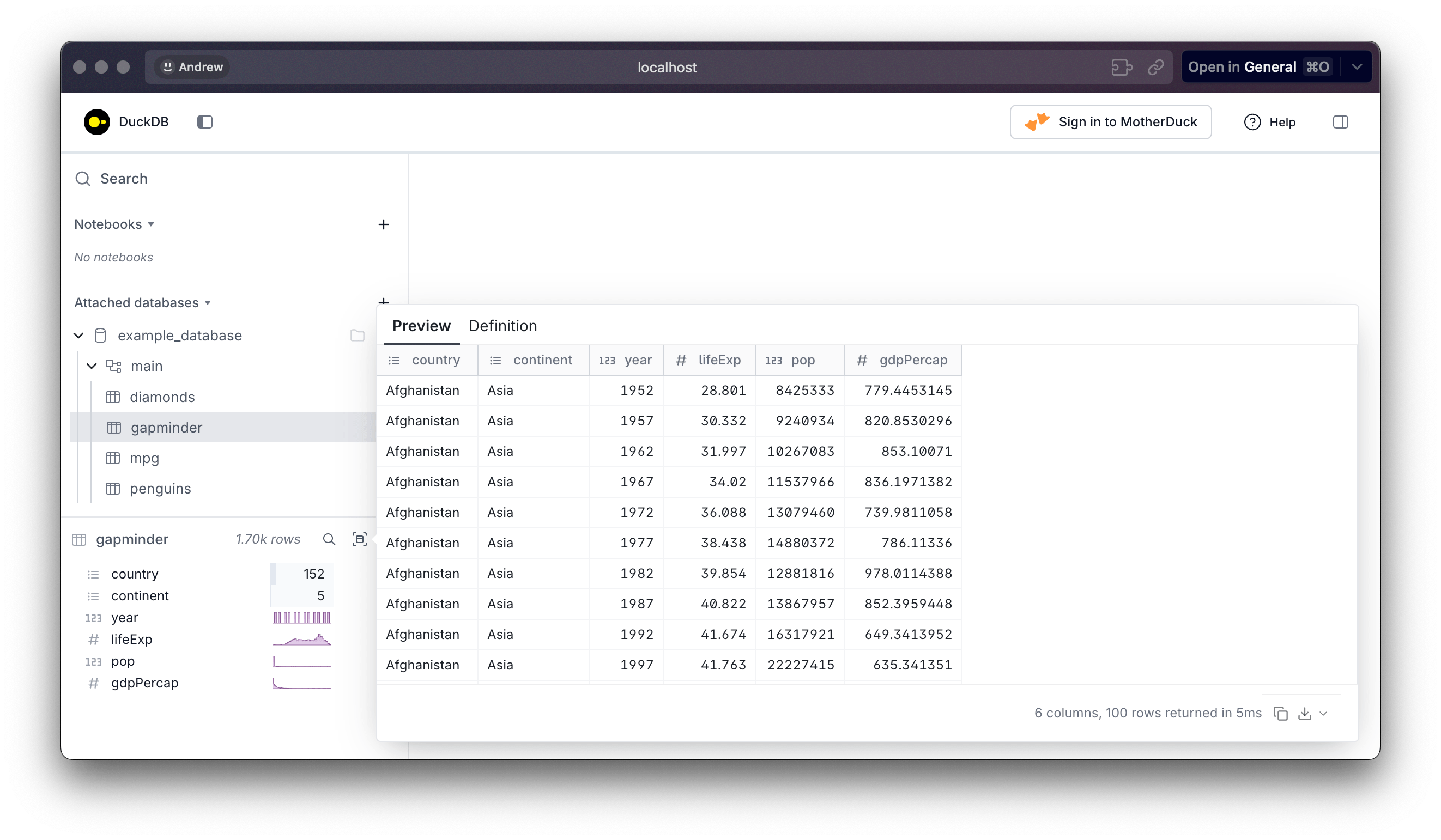
This all works fine for remembering what’s in the database, BUT it’s always a bit of a hassle because you can’t run commands from R as long as the the UI is open. You can only connect to a database once, so you’ll get this error:
con <- DBI::dbConnect(duckdb::duckdb(), db_file)
#> Error in `.local()`:
#> ! rapi_startup: Failed to open database: {"exception_type":"IO","exception_message":"Could not
#> set lock on file \"example_database.duckdb\": Conflicting lock is held in /opt/homebrew/Cellar/
#> duckdb/1.2.2/bin/duckdb (PID 48019) by user andrew.
#> See also https://duckdb.org/docs/stable/connect/concurrency","errno":"35"}When you’re done with the browser UI, you need to type .quit from the DuckDB terminal to close the connection:
❯ duckdb -ui example_database.duckdb
┌──────────────────────────────────────┐
│ result │
│ varchar │
├──────────────────────────────────────┤
│ UI started at http://localhost:4213/ │
└──────────────────────────────────────┘
v1.2.2 7c039464e4
Enter ".help" for usage hints.
D .quitSo you have to go back to the terminal, run .quit, and then go back to R and try again. If you want to check what’s in the database later, you have to disconnect from R, go to the terminal, run duckdb -ui, explore around in the browser, run .quit again, and go back to R.
DuckDB, {connections}, and the magical Connections Pane
There’s a better way to do this though! RStudio and Positron both have a Connections Pane that lets you save and explore database connections, and it magically fixes the problem with concurrent connections to a database so you can explore a database from inside your R session without needing to remember to turn off the in-browser GUI.
To make this work, you need to use the {connections} package, which is mostly a wrapper around standard {DBI} connection functions.
If you connect to the database with {connections}…
# Regular DBI way---don't do this
# con <- DBI::dbConnect(duckdb::duckdb(), db_file)
# Fancy {connections} way---do this. It connects *and* adds the connection to the sidebar
library(connections)
con <- connection_open(duckdb::duckdb(), db_file)…you’ll automatically see a new connection in Positron’s Connections Pane. Click on the little → arrow icon to open it:
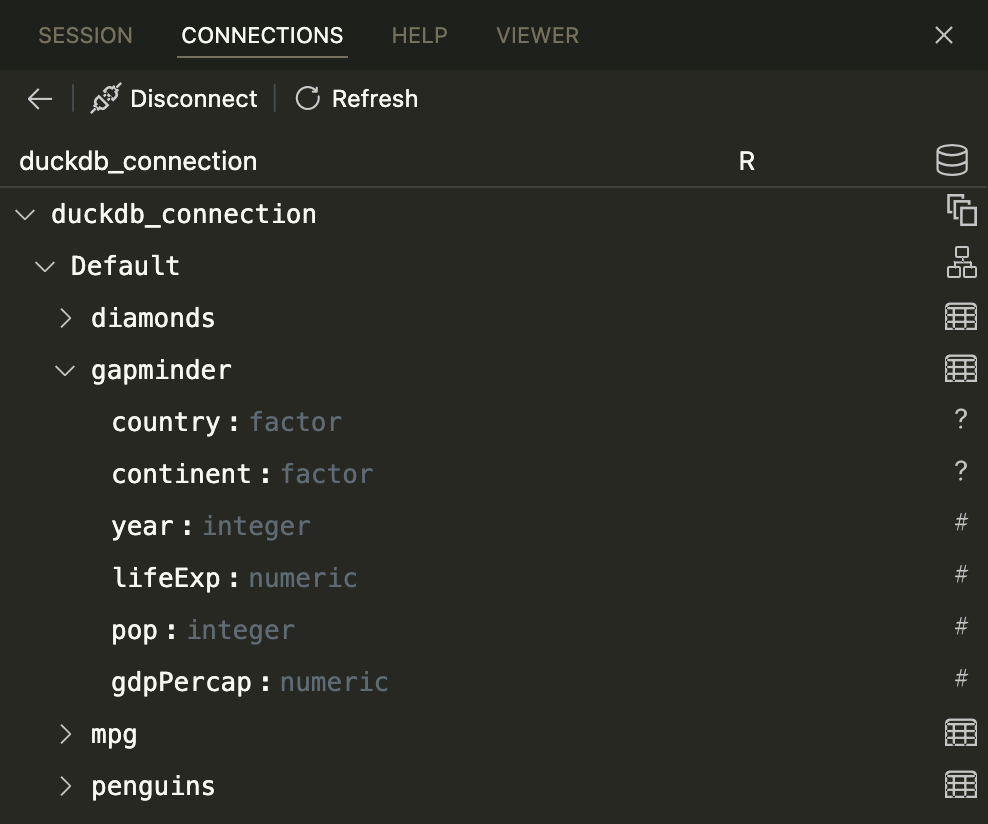
…and you’ll see all the tables inside!
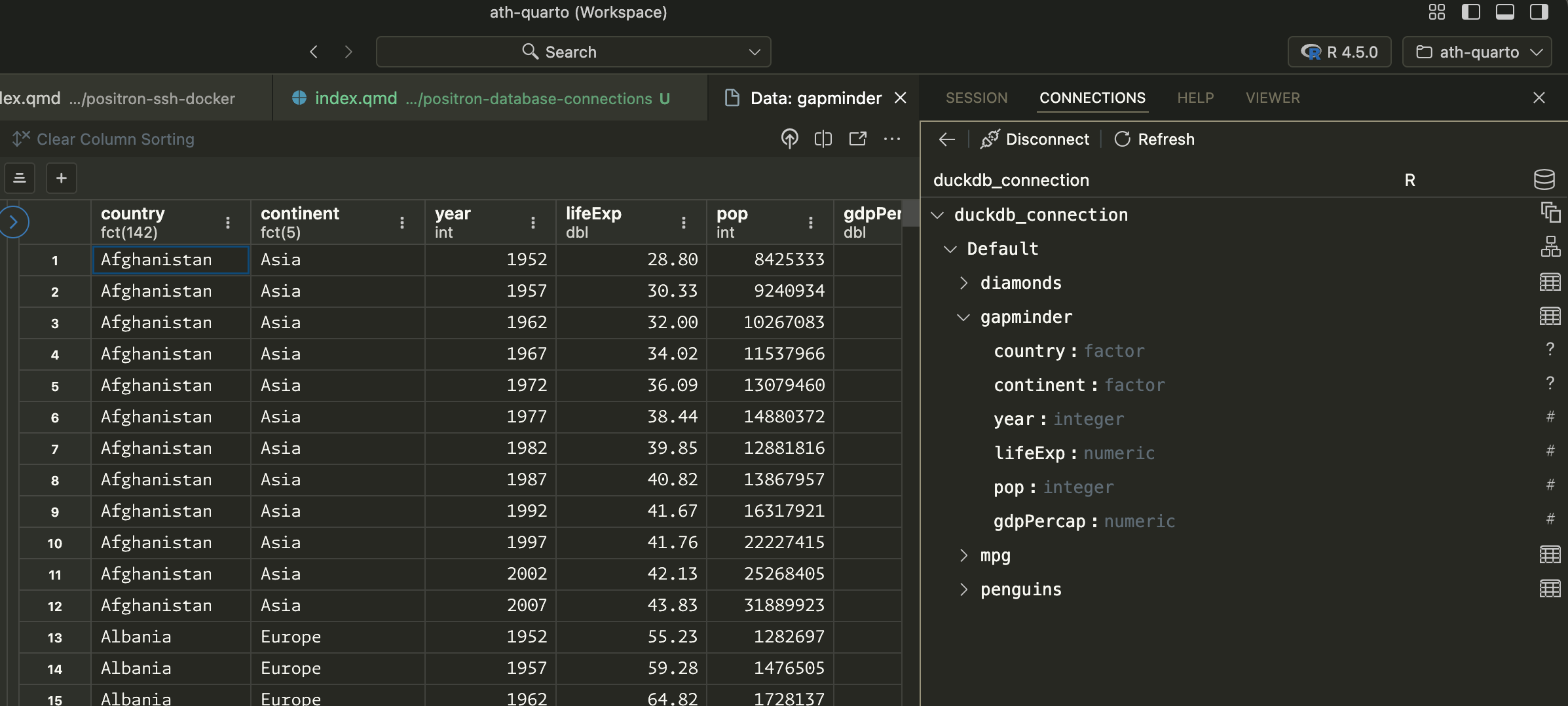
You can explore the data with Positron’s Data Explorer if you click on the little table icon:
And you can work with the database with R too, without running into concurrency issues:
# Get stuff out of the database
tbl(con, I("penguins")) |>
filter(species == "Adelie") |>
collect()
## # A tibble: 152 × 8
## species island bill_len bill_dep flipper_len body_mass sex year
## <fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
## 1 Adelie Torgersen 39.1 18.7 181 3750 male 2007
## 2 Adelie Torgersen 39.5 17.4 186 3800 female 2007
## 3 Adelie Torgersen 40.3 18 195 3250 female 2007
## 4 Adelie Torgersen NA NA NA NA <NA> 2007
## 5 Adelie Torgersen 36.7 19.3 193 3450 female 2007
## 6 Adelie Torgersen 39.3 20.6 190 3650 male 2007
## 7 Adelie Torgersen 38.9 17.8 181 3625 female 2007
## 8 Adelie Torgersen 39.2 19.6 195 4675 male 2007
## 9 Adelie Torgersen 34.1 18.1 193 3475 <NA> 2007
## 10 Adelie Torgersen 42 20.2 190 4250 <NA> 2007
## # ℹ 142 more rowsWhen you’re all done, disconnect from the database:
# Regular DBI way---don't do this
# DBI::dbDisconnect(con)
# Fancy {connections} way---do this
connections::connection_close(con)Bonus: Better support for DuckDB in the Connections Pane
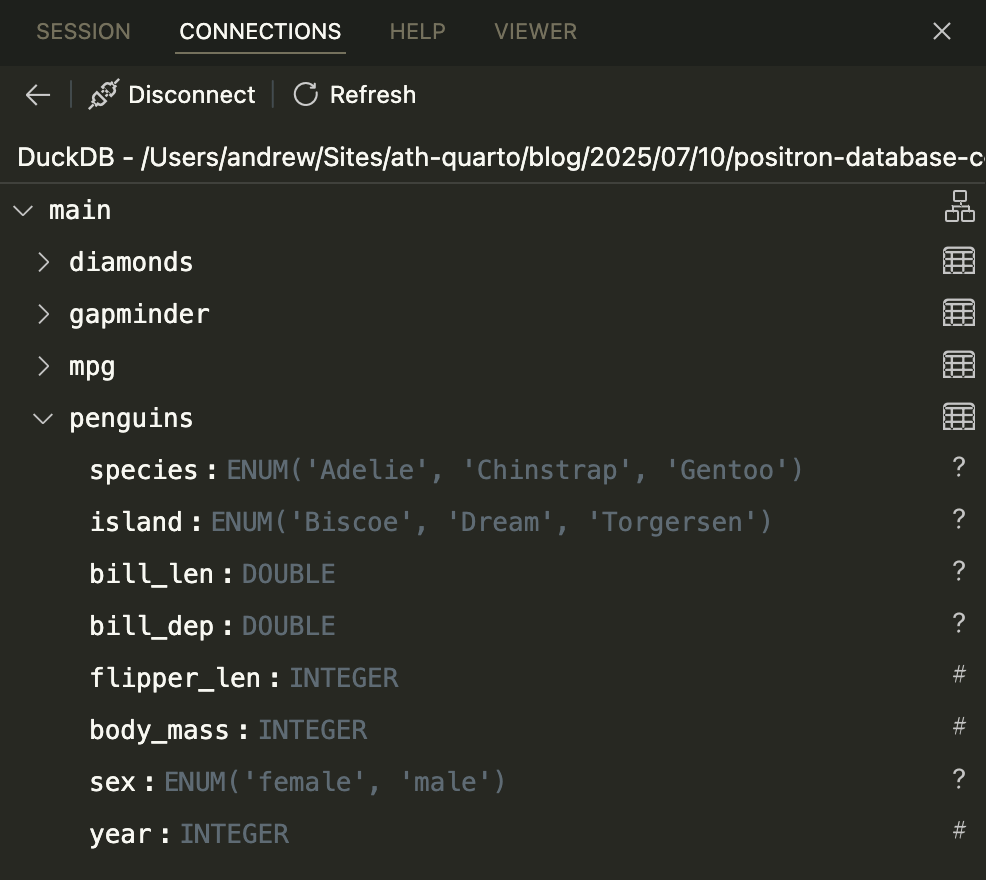
That Connections Pane connection is neat, but it’s missing a little bit of extra metadata (like the location of the file), and it shows the columns using generic output, not with DuckDB-specific column types.
DuckDB has fancier support for the Connections Pane, but for whatever reason, as of July 2025, it’s not enabled by default. If you set an option before connecting, though, you’ll get a prettier DuckDB-specific connection with a nice icon, the file location, and more specific column types.
options("duckdb.enable_rstudio_connection_pane" = TRUE)
con <- connections::connection_open(duckdb::duckdb(), db_file)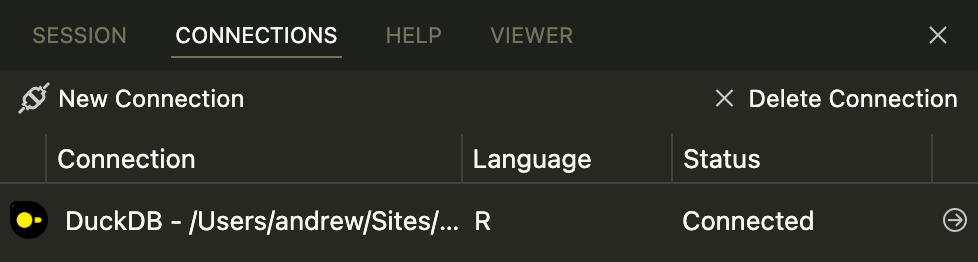
The whole game
Here’s what the overall process looks like for connecting to a database, adding data to it, querying it, closing the connection, and making a plot:
library(tidyverse)
# Use nicer DuckDB Connections Pane features
options("duckdb.enable_rstudio_connection_pane" = TRUE)
# Connect to an in-memory database, just for illustration
con <- connections::connection_open(duckdb::duckdb(), ":memory:")
# Add stuff to it
copy_to(
con,
gapminder::gapminder,
name = "gapminder",
overwrite = TRUE,
temporary = FALSE
)
## # Source: table<gapminder> [?? x 6]
## # Database: DuckDB v1.3.2 [root@Darwin 24.5.0:R 4.5.0/:memory:]
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Afghanistan Asia 1952 28.8 8425333 779.
## 2 Afghanistan Asia 1957 30.3 9240934 821.
## 3 Afghanistan Asia 1962 32.0 10267083 853.
## 4 Afghanistan Asia 1967 34.0 11537966 836.
## 5 Afghanistan Asia 1972 36.1 13079460 740.
## 6 Afghanistan Asia 1977 38.4 14880372 786.
## 7 Afghanistan Asia 1982 39.9 12881816 978.
## 8 Afghanistan Asia 1987 40.8 13867957 852.
## 9 Afghanistan Asia 1992 41.7 16317921 649.
## 10 Afghanistan Asia 1997 41.8 22227415 635.
## # ℹ more rows
# Get stuff out of it
gapminder_2007 <- tbl(con, I("gapminder")) |>
filter(year == 2007) |>
collect()
# All done
connections::connection_close(con)
# Make a pretty plot, just for fun
ggplot(gapminder_2007, aes(x = gdpPercap, y = lifeExp)) +
geom_point(aes(color = continent)) +
scale_x_log10(labels = scales::label_dollar(accuracy = 1)) +
scale_color_brewer(palette = "Set1") +
labs(
x = "GDP per capita",
y = "Life expectancy",
color = NULL,
title = "This data came from a DuckDB database!"
) +
theme_minimal(base_family = "Roboto Condensed")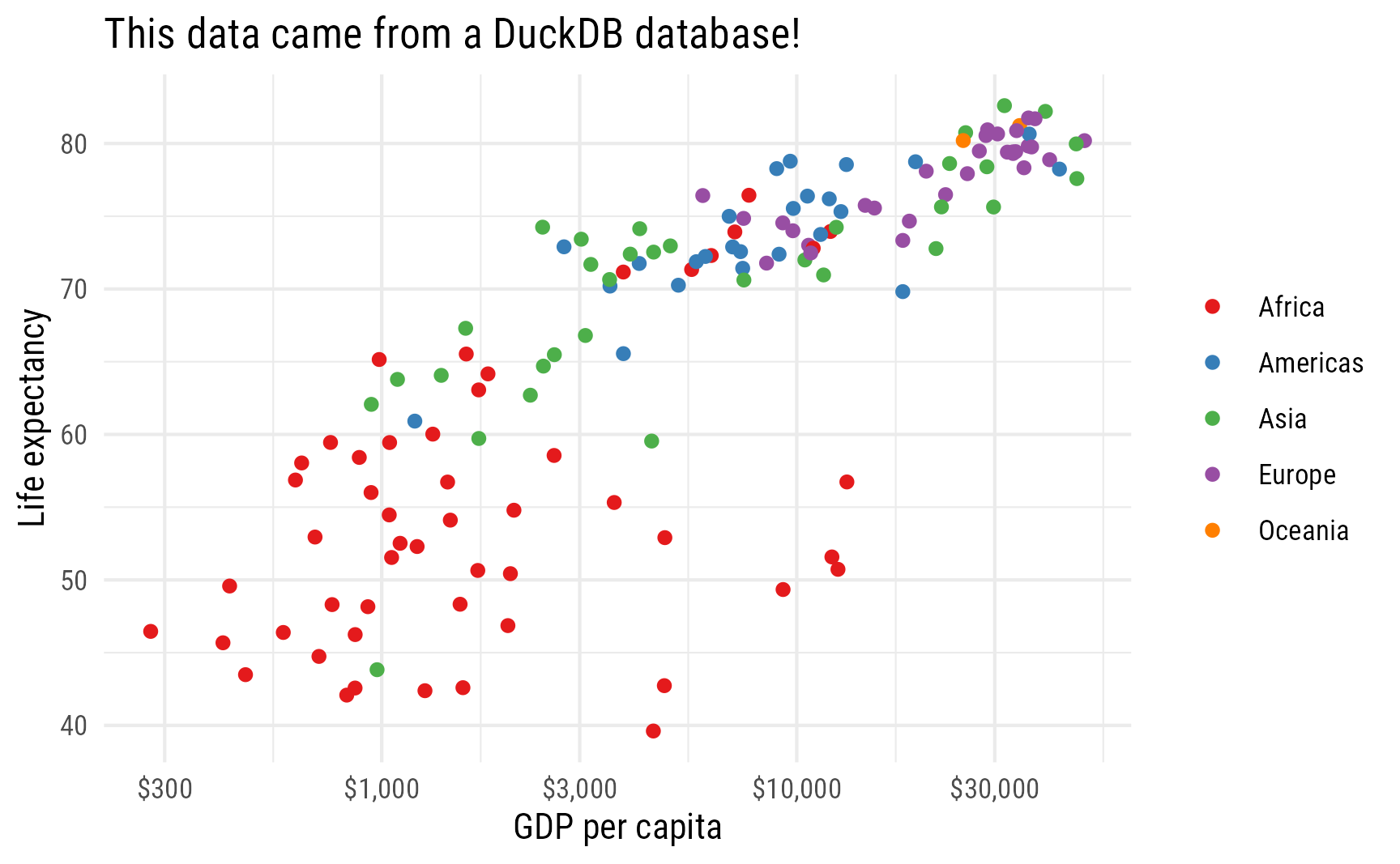
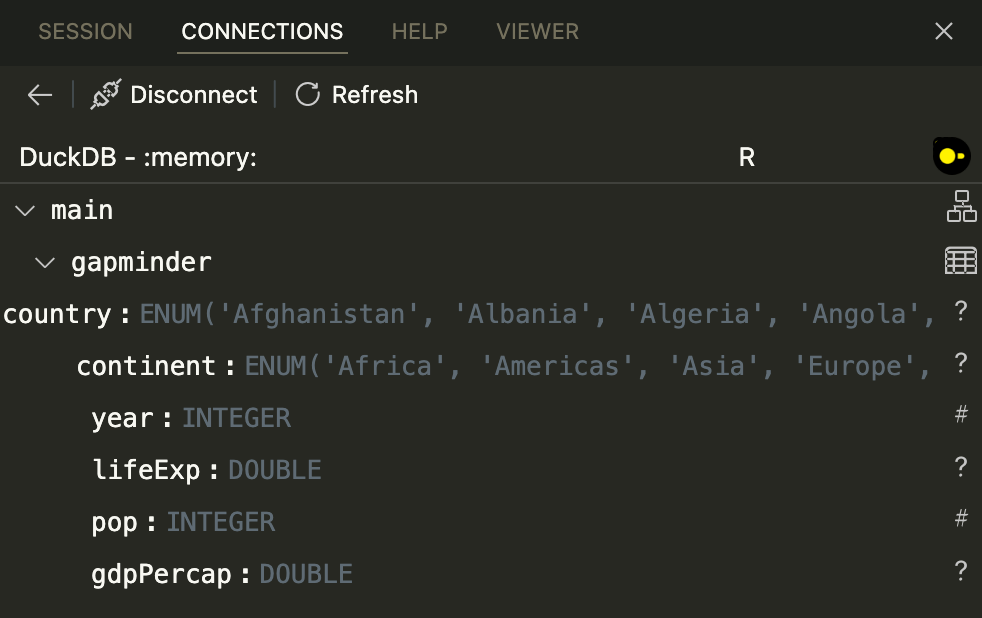
It works, and it shows the in-memory database in the Connections Pane!
Citation
@online{heiss2025,
author = {Heiss, Andrew},
title = {How to Use {Positron’s} {Connections} {Pane} with {DuckDB}},
date = {2025-07-10},
url = {https://www.andrewheiss.com/blog/2025/07/10/positron-database-connections/},
doi = {10.59350/w37d8-vj489},
langid = {en}
}