I’ve long been a proponent of making quantitative research reproducible. It’s the main reason I do all my scientific writing in Quarto—I can mix code and text in the same document so I don’t need to copy/paste numbers, tables, and figures from some statistical program into a word processor. Everything automatically ends up one compiled document based on the most current data.
There’s a growing trend where academic journals require that they be able to reproduce your results on their end before publication. For most articles in these journals, this involves having a PhD student run a bunch of Stata .do files, look through the logs and output, and ensure all the numbers and figures match what’s in the manuscript.
With a couple of my recent papers where the journal required this, I handed them a {targets} pipeline. The research assistant ran one line of R code (targets::tar_make()) to compile all the raw data, clean it, run several {brms} Bayesian models, and generate HTML, Word, and PDF versions of the manuscript through Quarto with all the latest values and tables and figures. Overkill, yes. But it’s so cool.
But getting the computing environment set up for this to work is difficult. Despite progress with {renv}, it’s still fairly tricky to get project-specific environments with both R and the project packages installed at specific versions. Unless you use something like rig, R by default uses the same installation systemwide, so once you upgrade from from something like R 4.4.3 to 4.5.0, you’ll use 4.5.0 for everything. {renv} lets you install packages at specific versions within folders, but even then it can often run into dependency hell if the lockfile gets out of sync or packages are removed from CRAN or GitHub (though Posit Package Manager aleviates a lot of this). It’s even trickier if you’re using other tools beyond R in your analysis—try installing Stan and CmdStanR in a cross platform way, with all the extra gcc compilers and Makefile settings!
In one recent project, I wrote up detailed instructions for installing the computing environment with {renv}, but it took a ton of back-and-forth emailing with the editorial office because their RA used an older version of Windows and they couldn’t get several parts of the environment installed.
To make life easier for everyone, I decided the best option for maximum cross platform reproducibility was to essentially give the journal’s RA a whole virtual computer with everything installed in it, all through the magic of Docker. Anyone can reproduce the results by following these instructions—in short, this uses Docker Compose to (1) build a Linux computer with R locked at version 4.4.1, thanks to the Rocker Project, (2) install all the version-locked R packages, which takes ≈15–20 minutes, (3) run a {targets} pipeline, which takes ≈20 minutes, and (4) spit out a final HTML/PDF/Word version of the manuscript and an accompanying analysis notebook.
Again, probably (definitely) overkill. But the RA at that journal was able to reproduce everything right away with Docker, and RAs at other journals have since done it flawlessly without a ton of back-and-forth troubleshooting.
But this post isn’t about that. Installing and configuring everything to get Docker Compose + rocker/tidyverse + {renv} + CmdStanR + Quarto working nicely is beyond the scope of this. It’s (unfortunately) left as an exercise to the reader. But I have heavily commented and documented examples if you want to try it! See the Dockerfile and docker-compose.yml here and the README for the project for a complete example.
This post is a lot simpler. It’s about how to actually use R within these containers and how the advent of Positron makes it way easier to write code in a regular IDE instead of inside a browser-based instance of RStudio.
How to use Docker-based R normally
Many (most?) of the Rocker R images come with RStudio Server built-in so that you can work with the container-based version of R directly from your browser.
Download this repository for a quick little example. It runs this Docker Compose file, which grabs the R 4.5.0 rocker/tidyverse, makes and RStudio Server available at http://localhost:8787, and mounts the main project folder into the container so that it’s mirrored and any file changes are persistent.
docker-compose-basic.yml
services:
rstudio:
platform: linux/amd64 # Need to include this when building on Silicon macOS
image: rocker/tidyverse:4.5.0
ports:
- "8787:8787" # Make localhost:8787 in the container accessible as localhost:8787 on the computer
environment:
- DISABLE_AUTH=true # Allow password-less login to RStudio server instance
volumes:
- ./project:/home/rstudio/project # Mount the local project folder into the containerHere’s how to get started:
- Install Docker Desktop on your computer (instructions for macOS or Windows).
- Install either (1) Positron and the Container Tools extension or (2) Visual Studio Code and its Container Tools extension. This is optional, since you can do all this Docker work with the terminal, but it makes working with Docker and Docker Compose way easier.
- Open the example folder in Positron or Visual Studio Code. In the File Explorer Pane, right click on
docker-compose-basic.yamland choose “Compose Up”. Or alternatively, using a terminal, navigate to the example folder and rundocker compose -f docker-compose-basic.yml up -d - Wait for the container to build. It goes pretty quickly, depending on your internet speed.
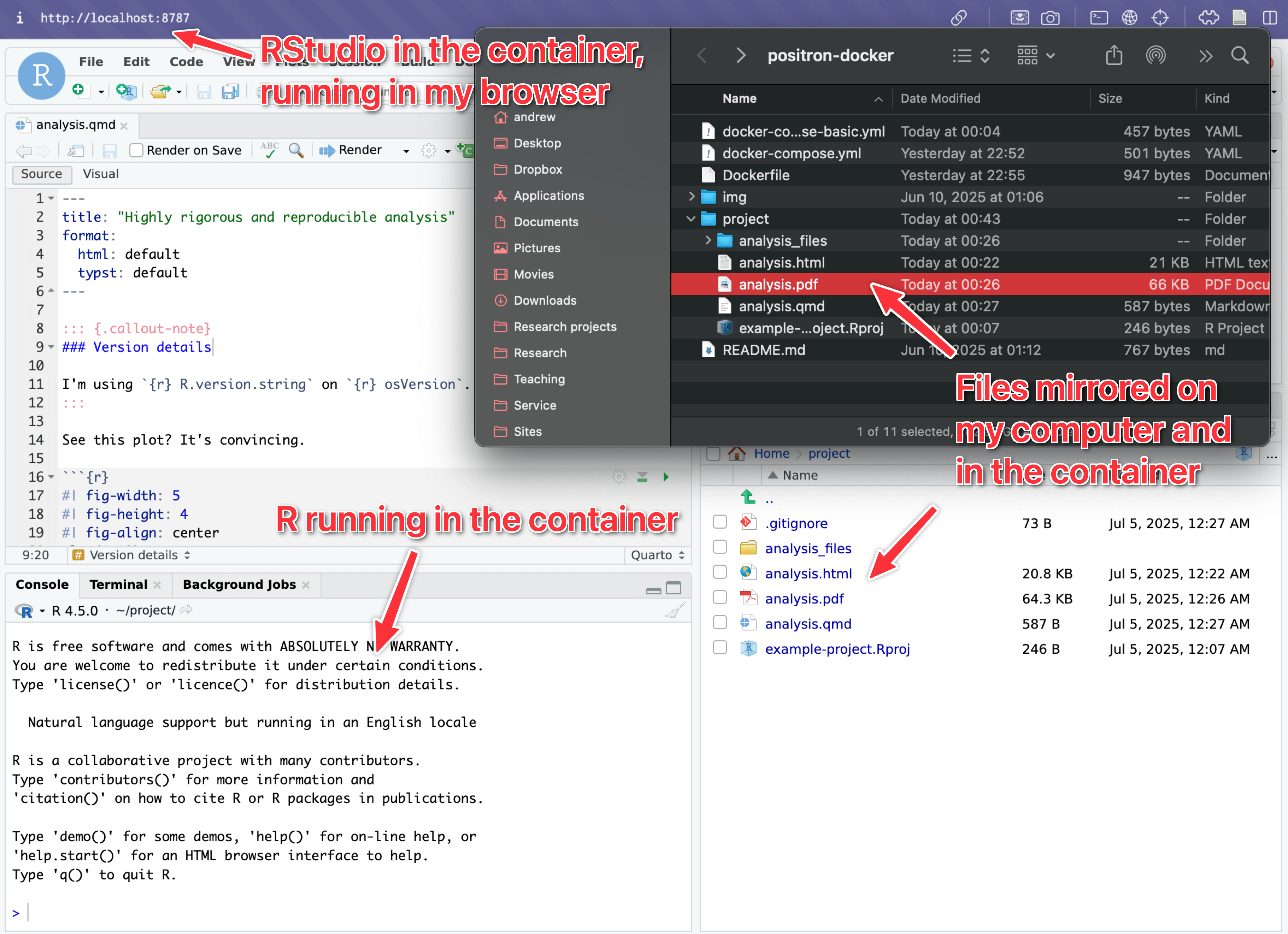
- Go to http://localhost:8787 in your browser and you’ll have RStudio in your browser!
- Open
project/and click onexample-project.Rprojto open an RStudio Project for the example R code. - Render
analysis.qmdto run the analysis.
The project/ folder in the example will automatically be mounted into the Docker container, so any edits you make while in the container will be saved on your local computer. If you render the analysis file, it’ll create an HTML file or Typst-based PDF, and those will appear on your local computer too because of the mounted folder
This process is super neat. You can access the container-based R from your browser and run and edit everything interactively there, and all changes and output will automatically show up on your local computer.
The main downside to this, though, is that you have to use RStudio in the browser. It doesn’t know about any custom settings and configuration you have in your regular RStudio installation on your computer. If you like to work in a dark theme with a ligature-based font like Fira Code and you have a bunch of custom keyboard shortcuts set up, or custom panel arrangements, or anything else, none of those changes will appear in the browser version because it’s a completely different version of RStudio. You can change settings for RStudio in the browser version, but unless you change the Docker Compose file to make it so that RStudio’s settings files/folders are mounted to your computer, any changes you make are ephemeral and will disappear the next time you run the Docker image. Also, your browser’s keyboard shortcuts will conflict with RStudio’s keyboard shortcuts. Like in Arc, ⌘S is used to toggle the tab sidebar. If I use it to save a file in RStudio, it’ll complain. And—maybe most annoyingly—if you use the ⌘W keyboard shortcut to close a file tab inside RStudio, it’ll actually close the whole browser tab.
Doing actual development in the container/browser version of RStudio is (1) important, since you get to use the version-locked packages and R installation, but (2) annoying because it feels like using someone else’s computer without all your tweaks and adjustments. Because of this, in practice, for projects where I care about exact replicability with version-locked packages and dependencies, I generally do all my coding and writing and analysis on my local computer and then try running it in the container at the end.
It would be really neat if there was some way to point RStudio or Positron to the container-based version of R and its package library so that I can use my regular local IDE, but backed by the container environment, but there’s no way to do that—both RStudio and Positron can only run local R installations.
How to use Docker-based R from Positron
But with Positron, there’s now (kind of) a way! Positron has native support for remote SSH sessions, where a whole local Positron window actually runs on a remote server. I use this (and Visual Studio Code’s SSH support) all the time for accessing files on my web server and doing anything else SSH-related.
The trick here is to use Positron to SSH into the container rather than into some remote machine.
The Rocker R images don’t have SSH enabled—or even installed—so we have to do a little extra work to do that. The docker-compose.yml and Dockerfile files in the example repository show all the changes that need to happen.
In short, we use a custom Dockerfile to pull rocker/tidyverse at R 4.5.0, and then we install and configure an SSH server on it. We can either set it up to require a password to connect (which is annoying) or use password-less public key authentication (which is not annoying once you have a public key set up).
Before trying this, make sure you have a public SSH key set up and accessible at ~/.ssh on your computer. Here’s a little guide.
Here’s the updated docker-compose.yml and the new Dockerfile:
docker-compose.yml
services:
tidyverse:
platform: linux/amd64 # Need to include this when building on M1 macOS
build:
# Build the Dockerfile
context: "."
dockerfile: "Dockerfile"
ports:
- "8787:8787"
- "2222:22" # localhost:2222 on the computer maps to localhost:22 in the container
environment:
- DISABLE_AUTH=true # Allow password-less login to RStudio server instance
volumes:
- ./project:/home/rstudio/project
# Mount the local public key into the container to SSH into it passwordlessly
- ~/.ssh/id_rsa.pub:/home/rstudio/.ssh/authorized_keys:ro Dockerfile
FROM rocker/tidyverse:4.5.0
# Install SSH
RUN apt-get update && \
apt-get install -y --no-install-recommends openssh-server sudo && \
rm -rf /var/lib/apt/lists/*
# Set up SSH
RUN mkdir -p /var/run/sshd && \
echo "PasswordAuthentication no" >> /etc/ssh/sshd_config && \
echo "PermitRootLogin no" >> /etc/ssh/sshd_config && \
echo "PubkeyAuthentication yes" >> /etc/ssh/sshd_config
# Password-less sudo in case things need to be installed from the terminal
# I have no idea what the sudo password is for this Docker container ¯\_(ツ)_/¯
RUN echo "rstudio ALL=(ALL) NOPASSWD:ALL" >> /etc/sudoers
# Make sure there's a .ssh folder with the right permissions. The
# local id_rsa.pub gets mounted to here through Docker Compose
RUN mkdir -p /home/rstudio/.ssh && \
chown rstudio:rstudio /home/rstudio/.ssh && \
chmod 700 /home/rstudio/.ssh
EXPOSE 22 8787
# Start both SSH and RStudio
CMD service ssh start && /initRight click on docker-compose.yml in the File Explorer Pane in Positron or Visual Studio Code and choose “Compose Up”, or run docker compose -f docker-compose.yml up -d in a terminal.
Once it’s done building, the container will still have an RStudio Server instance available at http://localhost:8787, and it will also have an SSH server exposed at port 2222. You can connect to it from the terminal on your computer with:
ssh rstudio@localhost -p 2222Or you can set up a saved connection (which Positron will be able to read) by adding this entry to ~/.ssh/config:
~/.ssh/config
Host positron-docker
HostName localhost
User rstudio
Port 2222With that shortcut, you can connect like this from the terminal:
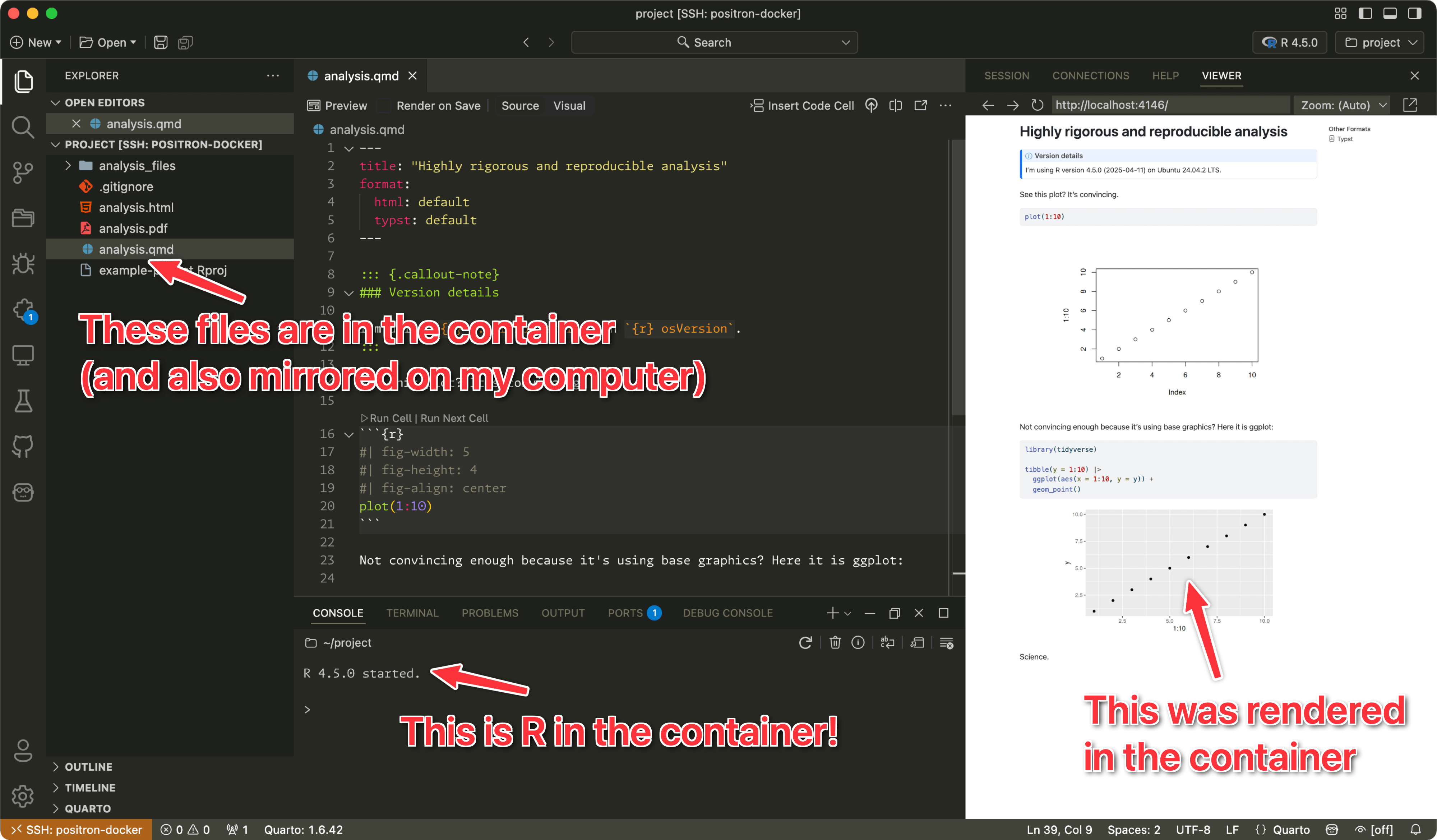
ssh positron-dockerEven better, once you add the connection to ~/.ssh/config, you can use Positron’s Remote Explorer to connect to the not-actually-remote remote SSH server running inside the container:
This gives you a(n almost) fully armed and operational Positron session that uses the version-locked installation of R inside the container. All your settings, preferences, themes, fonts, and keyboard shortcuts work here. It’ll feel (almost) just like a local Positron instance. The only thing that doesn’t transfer over to the containerized version of R (or any remote SSH instance) is extensions—Positron/Visual Studio Code will not automatically install your local extensions into the remote computer.
This makes it way easier to develop directly in the container. You get all your regular settings and you don’t get behavior that conflicts with the browser.
Citation
@online{heiss2025,
author = {Heiss, Andrew},
title = {Use {Positron} to Run {R} Inside a {Docker} Image Through
{SSH}},
date = {2025-07-05},
url = {https://www.andrewheiss.com/blog/2025/07/05/positron-ssh-docker/},
doi = {10.59350/fredm-56671},
langid = {en}
}